Symbiotic and dysbiotic oral microbiota: from health status to pathologies of the oral cavity.
UR D-258, Microbes, Evolution, Phylogénie et Infection (MEPHI)
Aix-Marseille Université (AMU)
Team leaders :
TERRER Elodie (PU-PH AMU/AP-HM)
MONNET-CORTI Virginie (PU-PH, AMU/AP-HM)
Members
ABOUDHARAM Gérard
ANTEZACK Angéline
DRANCOURT Michel
GRINE Ghiles
PILLIOL Virginie
TASSERY Hervé
Repertoire of the microbiota of the oral cavity and the periodontium: Isolate, cultivate, identify, characterize and keep in collection the newly isolated microorganisms.
New discoveries of microorganisms using microbiology techniques, microbial culture, fluorescence (FISH) and sequencing (MALDITOF). This is an important axis of our unit and the last four years have been marked in particular by the continuation of our efforts to describe the oral microbial repertoire by culturomics, in particular ancient microorganisms such as archaea, the discovery of the 1st nanoarchaea in humans, fastidious bacteria and to discover new species. Establishing the repertoire of microorganisms forming the oral microbiota is useful for understanding the health and diseases of the oral cavity (Belda-Ferre, 2012). The modern oral microbiota is established by analyzes of dental plaque, dental calculus and saliva, while the ancient oral microbiota can be reconstructed by analyzes based on the examination of ancient dental calculus. Ancient tartar has been recognized as the most informative source for paleomicrobiology (Adler, 2013; Warinner, 2014 and 2015).
Clinical archeology (methanogens, halophiles, nanoarchaea)
The archaea constitute one of the four domains of life alongside viruses, bacteria and eukaryotes (Boyer, 2010). These microorganisms, initially discovered in extreme environmental conditions, emerge as components of the oral, intestinal, vaginal mucosa and skin in humans (Probst, 2013; Nguyen-Hieu, 2013; Kulik, 2001; Lepp, 2004). The abundance and diversity of archaea in the oral cavity have mainly been studied through the detection of specific DNA sequences and metagenomic studies (Nguyen-Hieu, 2013). Indeed, methanogenic archaea are strict anaerobic prokaryotes whose culture conditions remain tedious and poorly known. Certain methanogenic archaea would be involved in periodontitis (Nguyen-Hieu, 2013; Bringuier, 2013).
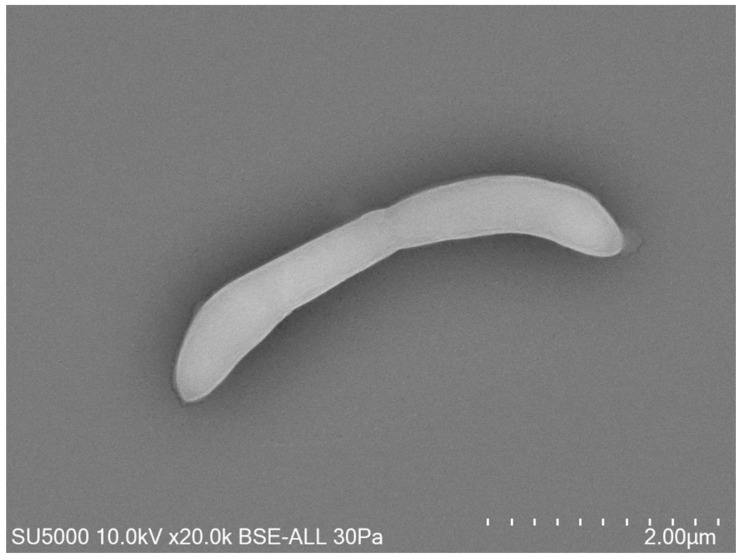
In recent years, we have been able to expand the repertoire of microorganisms in the oral cavity and in particular archaea since we have identified via saliva samples, Methanomassiliia massiliensis associated with a spirochete Massiliisphaerochaeta oralis (Guindo et al. 2022). From infected pulps we identified and cultured Methanobrevibacter oralis, Methanobrevibacter smithii, Methanobrevibacter massiliense (1/9, 11%) ass. Pyramidobacter piscolens (Pilliol et al., in progress). We also discovered this year the 1st nanoarchaea in humans (300-500 nm) in coculture, Nanopusillus massiliensis – Methanobrevibacter oralis / massilense thanks to dental plaque samples (Hassani et al. 2021). At present we are working on the genomic profile of antimicrobial resistance of nanoarchaea.
Improvement of materials and methods
We are continuing this work of identifying new pathogens via samples from the oral cavity (saliva, plaque, gingival fluid, calculus, dental pulp). To do this, we strive to improve the materials and methods used and to develop new detection methods and new strategies for culturing pathogens from the oral cavity. In this sense we have developed a new medium for the transport and culture of methanogens: the GG medium. This medium contains higher concentrations of acetate and formate than those of the SAB medium from which it is derived, but the GG medium, under a nitrogen atmosphere, makes it possible to overcome the dangerousness linked to the atmosphere concentrated in hydrogen of the SAB medium. This medium is the subject of a patent pending. Our development of an in vitro co-culture model for the culture and isolation of non-cultivable bacteria from the periodontium allowed us to describe a new species of Kingella, (Kingella bonacorsii) isolated from a patient undergoing periodontal maintenance (Antezack et al. al. 2021), a new species of Campylobacter (Campylobacter massiliensis sp. nov.), isolated from a patient with gingivitis (Antezack et al. 2021), a new species of Capnocytophaga (Capnocytophaga bilenii sp. Nov) isolated from a patient with gingivitis (Antezack et al. 2021), and a new species of Catonella (Catonella massiliensis sp. Nov).from a periodontal patient (Antezack et al. 2021).
Our research is focused on technology, the use and development of new tools are essential, for example the tabletop electron microscope.
Involvement of the oral microbiota in oral health and pathologies
Research focused on health and pathologies, in relation to the symbiotic and dysbiotic character of the oral microbiota:
– Relationship between the microbiota and periodontal health
– Relationship between the microbiota and periodontal diseases
– Relationship between the microbiota and peri-implant health
– Relationship between the microbiota and peri-implant diseases
– Relationship between the microbiota and pulp health
– Relationship between the microbiota and pulpal pathologies
– Determination of the role of archaea and nano archaea in infections of the oral cavity but also on the general state of health of the patient (archaea involved in infective endocarditis (Drancourt et al., 2021) or brain abscesses).
The work of our team aims to characterize the relationship between the oral microbiota and oral health and diseases, improve the prognosis of pathologies and the management of patients by implementing appropriate therapies according to the pathogen(s). detected. Comparison of protein profiles of saliva, dental plaque and gingival fluid in patients with and without periodontitis by MALDI-TOF mass spectrometry. 141 subjects with or without periodontitis were included. Based on the differential expression of protein peaks between the two groups, diagnostic decision trees were constructed for each sample to arrive at a diagnostic test with 100% specificity and sensitivity (95% CI = [ 0.91;1]) when 2 samples were tested (Antezack et al. 2019).
Microbiota of the oral cavity
Introduction
The oral microbiome is composed of a diverse community of bacteria, fungi, protozoa, viruses and archaea. The maintenance of a state of equilibrium between this community and the host ensures oral health. The disruption of this balance and the establishment of a dysbiotic microbial community are at the origin of oral pathologies, the most common of which are: carious disease and periodontal disease.
The advent of high throughput sequencing has led to a better appreciation of the true diversity of the oral microbiome and the latest version of the human oral microbiome database lists 775 microbial species. However, about 30% of these are, to date, not cultured and only known from 16S rRNA sequence data. This is the case for representatives of the phylum Saccharibacteria or certain eukaryotic viruses that have been identified in the oral cavity on the basis of molecular analysis and that seem to be associated with periodontal diseases. The development of co-culture methods has recently allowed the identification of the first members of Saccharibacteria. The various species of the oral microbiome associated with periodontal disease that have not yet been cultured are as likely to play a role in the aetiopathogenesis of periodontal disease as the cultured bacteria.
It is clear that the development of optimal culture media and the identification of the diversity of the oral microbiota has a major role to play in the study and understanding of periodontal disease. The reproducibility of these methods will allow us to extend the field of isolation and culture to other microbial species that were previously unculturable. Metagenomic or metaproteomic analyses have provided us with a great deal of information in recent years, but the isolation and culture of microorganisms is the only way to characterise them phenotypically and genotypically in an exhaustive manner and also to study their virulence potential. The various species of the oral microbiome associated with periodontal disease and not yet cultured are all as likely to play a role in the aetiopathogenesis of periodontal disease as the cultured bacteria. Thus, it is clear that the development of optimal culture media and the identification of the diversity of the oral microbiota has a major role in the study and understanding of periodontal disease.
It will be possible, after approval by a personal protection committee (CPP) and an ethics committee (CE), to collect samples from routine treatment residues (extracted teeth for which an indication for extraction has been given) or to collect samples of saliva, plaque and gingival fluid during treatment in the dentistry services led by the team directors and members of the whole H.U. team. Our previous work has made it possible to develop methods and tools for detecting and culturing microorganisms and to contribute to a better understanding of the diversity of the oral microbiota, and to describe new species isolated from patients treated in the periodontology department (Prof. Virginie Monnet-Corti) and the restorative dentistry and endodontics department (Prof. Elodie Terrer) (Annexe 1).
Strengths: The technical platform of the IHU Méditerranée Infection and our previous work allow us to carry out the analysis of samples with a mainly descriptive and secondarily comparative aim.
Opportunities: Thanks to our respective university hospital services, we have the possibility to conduct clinical studies including a large number of subjects with periodontal/pulpal diseases as well as subjects without these pathologies. The interest in determining the pathological or non-pathological role of the identified species in oral cavity diseases could lead to a patent and an industrial partnership to develop rapid, inexpensive and reliable kits to screen for a pathological and non-pathological state of the oral cavity tissues. Our work will be disseminated on social networks. This promotional work should be developed by integrating the socio-economic partners. We will develop our investment in the communication of scientific information to the general public.
Weaknesses: Lack of permanent research technicians and engineers
Threats: Culturomics in general and the cultivation (co-culture) of fastidious microbes in particular are time-consuming and require a high level of handling. They require full-time work with trained and competent staff.
Situation report on current microbiological cultivation in the world: approaches of our team and development of new strategies
The preparation and analysis of the samples will be carried out at the IHU, in the MEPHI laboratory. The samples will be cultured in different media and different atmospheric conditions will be tested in order to develop optimal co-culture models. The samples will be analysed by various molecular biology methods in order to detect the species of interest and study their genomes. Different imaging methods will be used on the samples and on the species isolated by the co-culture methods in order to make morphological observations, to evaluate their growth and viability, to study and characterise the species.
- Building up collections of cultivable microbes
- Improvement of procedures for the isolation of cultivable microbes
- Development of simple methods for the isolation of non-cultivable/miscultivable microbes
- Development of in vitro co-culture models for the culture and isolation of non-cultivated microbial species from the oral cavity.
- Measurement of the performance of co-culture models on the isolation and growth of non-cultivable species
- Isolation of new microbes by culture
Combining genomic and metagenomic data for the detection of new genotypes of oral cavity microbes
- Microbial metagenomics
- Analysis against local genomic library
- Targeted culture/PCR4.
Relationship between human oral cavity diseases and various microbiota
- To study the diversity of the oral microbiota, associated with periodontal disease, using co-culture, molecular biology and imaging methods
- To compare the composition of the microbiota and the distribution of species in subjects according to medical and periodontal/dental status (pathological or non-pathological).
- Identify species associated with periodontal disease.
To investigate the association between patient medical characteristics (gingival health or periodontal/peri-implant disease) and the species identified and cultured.e. To study the synergy between certain species within the healthy and pathological oral microbiota
Valorization of research
– Clinical valorization: development of identification and diagnostic tools, use of biomarkers allowing in particular the improvement of screening and diagnosis of pathologies of the oral cavity. Development, evaluation and marketing of one or more oral POCs which would allow, through self-saliva sampling, to carry out a rapid diagnosis of the pathogens of interest.
– Pedagogical valorization: obvious valorization of useful research for the purpose of updating our teaching concerning students and interns in health, but also science students.
Paleomicrobiology
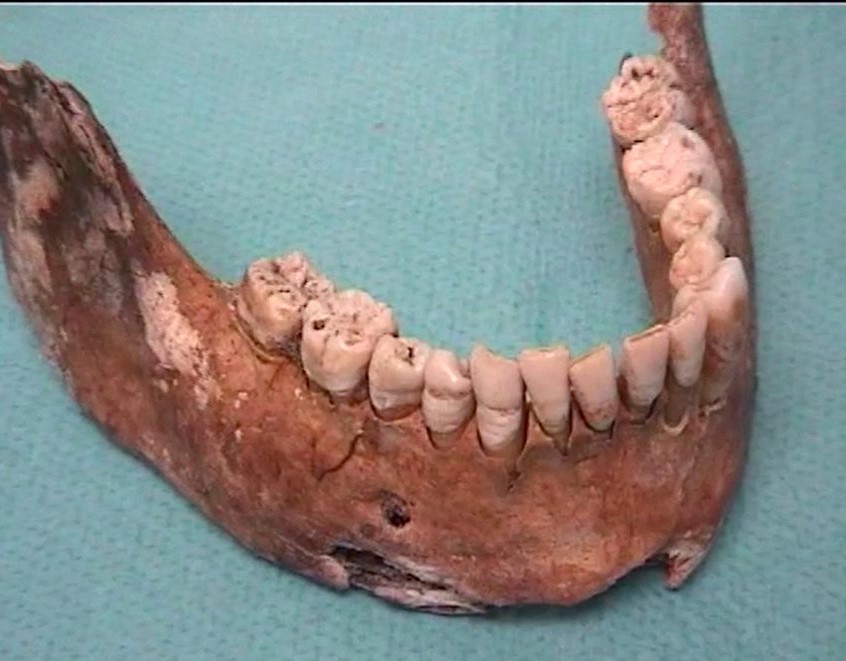
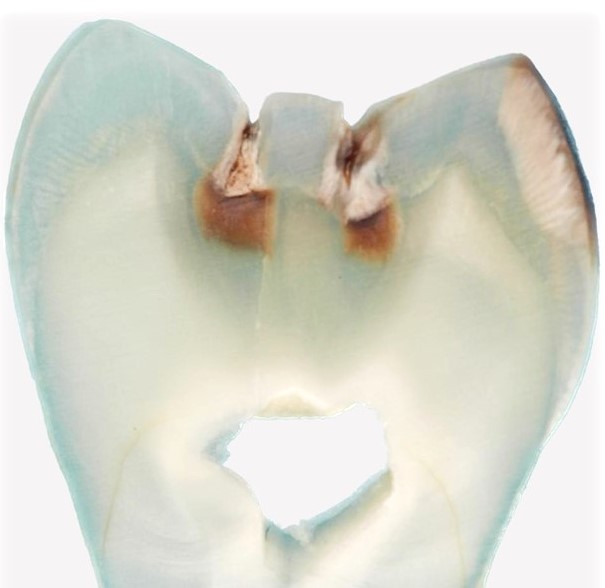
Paleomicrobiology is the study of the microbiota initially passed from old dental pulp. The dental pulp is a particularly appropriate tool to recover the bacteria responsible for the infection after bacteremia (Aboudharam, 2004). The use of dental pulp as a potential source of DNA represents a biological material of choice given the excellent results obtained (Pillay and Kramer, 1997). The quantities of DNA extracted from the pulp are small but nevertheless of high quality due to the preservation of the connective tissues inside the pulp chamber. Few studies focus on the use of the tooth for the search for pathogens and in particular the use of pulp DNA for the characterization of pathogens. Hence the originality of this approach, which consisted in detecting bacterial DNA from pulp DNA.
Contribution of molecular biology
The disadvantage of the molecular strategy by designating specific primers for the species sought is to answer only a single etiological question oriented most of the time by anthropologists. The use of a universal detection and identification tool such as the gene encoding 16S RNA appears useful.
The dental pulp is used by molecular biology technique for the detection of DNA of microorganisms. However, this pulp must be removed from the tooth before use; Dental pulp extirpation protocols require dental expertise for proper pulp recovery. In addition, dental pulp samples exposed to the laboratory environment may be subject to contamination detected by the 16S rDNA used for the universal detection of bacteria.
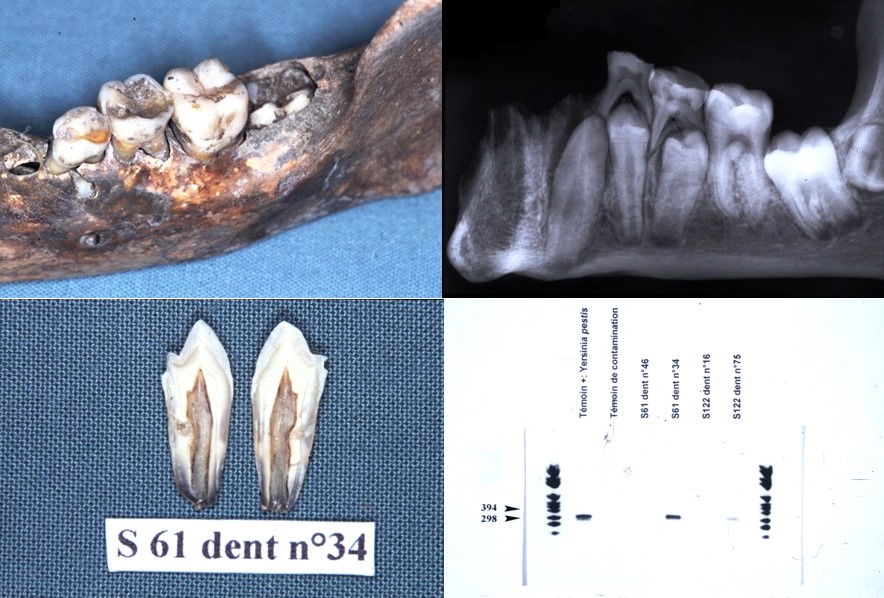
It is for these reasons that a new protocol has been developed using on the one hand a method of fixing the tooth in sterile resin, in order to extract the DNA directly in the pulp chamber and on the other hand by decontaminating PCR reagents by filtration and enzymatic digestion. This protocol prevents contamination and allows the detection of bacteria from the dental pulp responsible for colonization in the blood and periodontal tissues. Only 10% of samples contain 16S rDNA. It is a tool for the retrospective diagnosis of bacteremia using the universal detection of bacterial DNA in animals and humans, on old or contemporary teeth (Tran-Hung, 2007).
The evolution of paleomicrobiology work and in particular the paleoproteomic work of the dental pulp represents another new and innovative approach for detecting fragments of ancient peptides of various pathogens from dental pulps of individuals who died several hundred years ago. This new orientation of paleomicrobiology work has provided additional information to that obtained previously (Barbieri, 2017).
Other orientations consisting in reproducing tests usually used on blood are being developed (miniaturization) on the dental pulp. These tests, paleoserology and paleocytology, should provide additional information and new perspectives.
Paleoserology
The study of past and contemporary oral microbiota has enabled us to develop specific paleomicrobiology techniques, in particular paleoserology from samples of old pulps example of coronavirus detected from pulps from 1914, (Oumarou et al. 2021); a posteriori diagnosis of dysglobulinaemia from an old pulp (Oumarou et al. 2022); outbreak of relapsing fever (Borrelia recurrentis) unmasked by microbial paleoserology, 16th century, France (Oumarou et al., 2022). We are in the process of extrapolating these ancient pulp paleocytology techniques to other types of ancient samples from the oral cavity such as dental calculus (coronavirus detection under Napoleon from calculus samples from 1809) (Merrouche et al., in progress). We are also developing techniques for paleo autoimmunochemistry of old dental pulp.
Paleoculture
Paleoculture techniques deserve special attention, the use of precise skills and methods indeed the old samples used represent a precious and rare historical material. In addition, the size and quantity of equipment are limited, the experimentation of the operator is essential in order to best preserve the samples and the chances of detecting pathogens, the role of the dental surgeon who has a better knowledge and expertise of the samples of dental origin is important here. We try to develop microbiology techniques adapted to the sample size such as miniaturized culturomics.
Paleomicrobiology has a certain diagnostic interest (a posteriori diagnosis), it allows the improvement of historical knowledge since it provides information on the history of diseases and sometimes allows to modify current knowledge (diagnosis of Clostridium tertium bacteremia from a pulp from 1914 (Meucci et al., 2022); Clostridium tetani bacteraemia from pulp from 1590 (Bouri et al., in progress).
Exploring the role of dental pulp stem cells (DPSC)
The study of the past oral microbiota also allows us to explore the possible role of dental pulp stem cells (DPSC) and the survival of microorganisms in the pulp example of internalization Bartonella quintana in the DPSC of old pulps (Oumarou et al., 2021). DPSCs, like phagocytes, can take up and internalize Bartonella quintana and this internalization is associated with expression of pro-inflammatory and anti-inflammatory genes. This activity does not affect the stem and mesenchymal cell properties of DPSCs. These results support the hypothesis that the dental pulp and in particular the DPSCs could constitute a sanctuary for chronic Bartonella quintana infection.
Research activities
Area 1: Describe the human microbiota using culturomics and understand the relationships between the oral microbiota and oral health and diseases: the first objective is to significantly increase the known microbial repertoire in the humans using large-scale culture methods and MALDI-TOF identification, as well as genome sequencing. The second objective is to study the relationships between the microbiota and oral diseases such as gingivitis, periodontitis, peri-implantitis, pulp pathologies and the involvement of archaea and nanoarchaea in these different pathologies. The analysis of the oral microbiota continues to contribute to an inventory of the respective flora, with the objective of finding markers of oral health.
Area 2: Exploration of a new field of human microbiota including fastidious microorganisms which are little or badly known, despite the fact that they constitute a large part of the human oral microbiota. We focused on co-culture with auxiliaries to discover new oral microbial genera and species.
Area 3: Research focused on technology and using new tools: the example of tabletop electron microscopy. We have worked in different areas of the microbiota by applying this technology to clinical microbiology. Using scanning electron microscopy, correlative microscopy method and immunofluorescence, we showed the presence of SARSlike particles in RT-qPCR SARS-CoV-2-positive saliva, dental plaque and gingival fluid samples (Belhaouari et al. 2022).
Area 4: Discovery of ancient and modern pathogens. Development of paleoserology and paleoculture.
Area 5: Explore the role of pulp stem cells (DPSC) in past infections (example of Bartonella quintana) and contemporary (examples of infective endocarditis, Oumarou et al., 2021).
Network
- Within our MEPHI structure: collaboration with Prof. Colson’s future team on viruses of the oral cavity: a PhD student will be assigned to work on the association of certain emerging viruses with periodontal disease (CPP in progress). Possible collaborations with different teams of our research unit, in particular with the future team of Prof. Million concerning the implication of oral dysbiosis in severe malnutrition of children in West Africa.
- With research structures of Aix Marseille University: multiple collaborations in progress with archaeologists with M. Signoli and C. Costedoat of the ADES laboratory. These collaborations allow us to obtain valuable samples (tartars and ancient teeth) useful in palaeomicrobiology.
- With national research laboratories: collaborations with the CHU Estaing (Pr Hennequin, Clermont Ferrand), on the interface between the texture of the alimentary bolus and the modification of the oral microbiota, with the hospital Pontchaillou (Pr Bonnaure-Mallet, Rennes) around various themes, one of which is the influence of tobacco on the oral flora (PHRC project in progress), with the CHU of Nice (Pr Vincent-Bugnas and Dr A. Doglio, MICORALIS laboratory UPR7354), and the University Hospital of Rennes (Pr Jeanne) within the project HERPARO, (PHRC in progress).
Conclusions
A recent study has provided updated estimates of the direct and indirect costs of oral diseases at global, regional and national levels. The direct costs of oral diseases were estimated using a systematic approach; the indirect costs were estimated using an approach developed by the World Health Organization’s Commission on Macroeconomics and Health and taking into account the 2015 values for gross domestic product and disability-adjusted life years from the Global Burden of Disease study. In this study, the direct costs of oral diseases were estimated at $356.80 billion and the indirect costs at $187.61 billion, for a total of $544.41 billion in global costs attributed to oral diseases in 2015.
After adjusting for purchasing power parity, the highest levels of dental expenditure per person were found for the high-income countries of North America, Australia, Western Europe, high-income Asia Pacific and East Asia; the highest levels of productivity losses per person were found for Western Europe, high-income countries of North America, high-income Asia Pacific and Central Europe.
From an economic point of view, improving the oral health of the population can be beneficial and could contribute to increasing the well-being of individuals, given the resources available. The development of screening and early diagnosis tools would enable future prevention and control programmes for these oral diseases. The knowledge of the pathogens involved in the various oral diseases would allow the best adaptation of therapeutics, and as a direct consequence an improvement of the screening, the evolution of these pathologies and the management of the patients, but also an undeniable public health issue.
Publications
References
- Antezack A, Boxberger M, Rolland C, Ben Kheder M, Monnet-Corti V, La Scola B. Isolation and characterization of Campylobacter massiliensis sp. nov., a novel Campylobacter species detected in a gingivitis subject. Int J Syst Evol Microbiol. 2021;71(10). doi: 10.1099/ijsem.0.005039.
- Antezack A, Boxberger M, Ben Kheder M, La Scola B, Monnet-Corti V. Selenomonas timonae sp. nov., a new bacterium isolated from a gingivitis subject. Int J Syst Evol Microbiol. 2021;71:005040. doi: 10.1099/ijsem.0.005040.
- Boxberger M, Antezack A, Magnien S, Cassir N, La Scola B. Complete genome and description of Corynebacterium incognita strain Marseille-Q3630T sp.nov., a new bacterium isolated from the blood of a 7-years-old girl. New Microbes New Infect. 2021;42:100893. doi: 10.1016/j.nmni.2021.100893.
- Antezack A, Boxberger M, Monnet-Corti V, La Scola B. Isolation and characterization of Capnocytophaga bilenii sp. nov., a novel Capnocytophaga species detected in a gingivitis subject. Pathogens. 2021; 10(5):547. https://doi.org/10.3390/pathogens10050547.
- Antezack A, Boxberger M, La Scola B, Monnet-Corti V. Isolation and description of Catonella massiliensis sp. nov., a novel Catonella species, isolated from a stable periodontitis subject. Pathogens. 2021;19;10(3):367. doi:10.3390/pathogens10030367.
- Antezack A, Boxberber M, Rolland C, Monnet-Corti V, La Scola B. Isolation and characterization of Kingella bonacorsii sp. nov., a novel Kingella species detected in a stable periodontitis subject. Pathogens. 2021; 10(2):240. doi: 10.3390/pathogens10020240
- Brahim Belhaouari DB, Baudoin J-P, Lagier J-C, Monnet-Corti V, La Scola B, Antezack A. Microscopic observations of SARS-CoV-2 like particles in different oral samples. Eur J Oral Sci. 2022;e12903.
- Boxberger M, Antezack A, Magnien S, Cassir N, La Scola B. Draft genome and description of Mixta mediterraneensis strain Marseille-Q2057T sp.nov., a new bacterium isolated from human healthy skin. New Microbes New Infect. 2021;40:100840. doi: 10.1016/j.nmni.2021.100840.
- Boxberger M, Antezack A, Magnien S, Cassir N, La Scola B. Draft genome and description of Corynebacterium haemomassiliense strain Marseille-Q3615T sp. nov., a new bacterium isolated from a 59-year-old man with chronic obstructive pulmonary disease symptoms. New Microbes New Infect. 2020;38:100801. doi: 10.1016/j.nmni.2020.100801.
- Meucci M, Costedoat C, Verna E, Adam F, Signoli M, Drancourt M, Beye M, Aboudharam G, Barbieri R. Whole genome sequence of bacteremic Clostridium tertium in a World War I soldier, 1914. Current Research in Microbial Sciences, Elsevier. 2022;p 100089.
- Grine G, Royer A, Terrer E, Diallo OO, Drancourt M, Aboudharam G. Tobacco Smoking Affects the Salivary Gram-Positive Bacterial Population. Front Public Health. 2019;7:196. doi: 10.3389/fpubh.2019.00196. eCollection 2019.
- Guindo CO, Terrer E, Chabrière E, Aboudharam G, Drancourt M, Grine G. Culture of salivary methanogens assisted by chemically produced hydrogen. Anaerobe. 2020;61:102128. doi: 10.1016/j.anaerobe.2019.102128. Epub 2019 Nov 20.
- Grine G, Terrer E, Boualam MA, Aboudharam G, Chaudet H, Ruimy R, Drancourt M. Tobacco-smoking-related prevalence of methanogens in the oral fluid microbiota. Sci Rep. 2018;8(1):9197. doi: 10.1038/s41598-018-27372-7.
- Hassani Y, Saad J, Terrer E, Aboudharam G, Giancarlo B, Silvestri F, Raoult D, Drancourt M, Grine G. Introducing clinical nanorachaeaology: Isolation by co-culture of Nanopusillus massiliensis sp. nov. Curr Res Microb Sci. 2021;3:100100. doi: 10.1016/j.crmicr.2021.100100. eCollection 2022.
- Oumarou Hama H, Hamada A, Aboudharam G, Ghigo E, Drancourt M. Human dental pulp stem cells: A sanctuary for relapsing Bartonella quintana. Microbial Pathogenesis. 2021;153:104797.
- Ramassy L, Oumarou Hama H, Costedoat C, Signoli M, Verna E, La Scola B, Aboudharam G, Barbieri R, Drancourt M. Paleoserology points to Coronavirus as possible causative pathogens of the ‘Russian flu.’ Microb Biotechnol. 2022;15:1943–1945.
- Oumarou Hama H, Barbieri R, Guirou J, Chenal T, Mayer A, Ardagna Y, Signoli M, Aboudharam G, Raoult D, Drancourt M. An outbreak of relapsing fever unmasked by microbial paleoserology, 16th century, France. Am J Phys Anthropol. 2020;173:784–789.